arviz.plot_posterior#
- arviz.plot_posterior(data, var_names=None, filter_vars=None, combine_dims=None, transform=None, coords=None, grid=None, figsize=None, textsize=None, hdi_prob=None, multimodal=False, skipna=False, round_to=None, point_estimate='auto', group='posterior', rope=None, ref_val=None, rope_color='C2', ref_val_color='C1', kind=None, bw='default', circular=False, bins=None, labeller=None, ax=None, backend=None, backend_kwargs=None, show=None, **kwargs)[source]#
Plot Posterior densities in the style of John K. Kruschke’s book.
- Parameters:
- data: obj
Any object that can be converted to an
arviz.InferenceDataobject. Refer to the documentation ofarviz.convert_to_dataset()for details- var_names: list of variable names
Variables to be plotted, two variables are required. Prefix the variables with
~when you want to exclude them from the plot.- filter_vars: {None, “like”, “regex”}, optional, default=None
If
None(default), interpret var_names as the real variables names. If “like”, interpret var_names as substrings of the real variables names. If “regex”, interpret var_names as regular expressions on the real variables names. A lapandas.filter.- combine_dims
set_likeofstr, optional List of dimensions to reduce. Defaults to reducing only the “chain” and “draw” dimensions. See the this section for usage examples.
- transform: callable
Function to transform data (defaults to None i.e.the identity function)
- coords: mapping, optional
Coordinates of var_names to be plotted. Passed to
xarray.Dataset.sel()- grid
tuple Number of rows and columns. Defaults to None, the rows and columns are automatically inferred.
- figsize: tuple
Figure size. If None it will be defined automatically.
- textsize: float
Text size scaling factor for labels, titles and lines. If None it will be autoscaled based on
figsize.- hdi_prob: float, optional
Plots highest density interval for chosen percentage of density. Use ‘hide’ to hide the highest density interval. Defaults to 0.94.
- multimodal: bool
If true (default) it may compute more than one credible interval if the distribution is multimodal and the modes are well separated.
- skipnabool
If true ignores nan values when computing the hdi and point estimates. Defaults to false.
- round_to: int, optional
Controls formatting of floats. Defaults to 2 or the integer part, whichever is bigger.
- point_estimate: Optional[str]
Plot point estimate per variable. Values should be ‘mean’, ‘median’, ‘mode’ or None. Defaults to ‘auto’ i.e. it falls back to default set in rcParams.
- group: str, optional
Specifies which InferenceData group should be plotted. Defaults to ‘posterior’.
- rope
list,tupleordictionaryof {str:tuplesorlists}, optional A dictionary of tuples with the lower and upper values of the Region Of Practical Equivalence. See this section for usage examples.
- ref_val: float or dictionary of floats
display the percentage below and above the values in ref_val. Must be None (default), a constant, a list or a dictionary like see an example below. If a list is provided, its length should match the number of variables.
- rope_color: str, optional
Specifies the color of ROPE and displayed percentage within ROPE
- ref_val_color: str, optional
Specifies the color of the displayed percentage
- kind: str
Type of plot to display (kde or hist) For discrete variables this argument is ignored and a histogram is always used. Defaults to rcParam
plot.density_kind- bw: float or str, optional
If numeric, indicates the bandwidth and must be positive. If str, indicates the method to estimate the bandwidth and must be one of “scott”, “silverman”, “isj” or “experimental” when
circularis False and “taylor” (for now) whencircularis True. Defaults to “default” which means “experimental” when variable is not circular and “taylor” when it is. Only works ifkind == kde.- circular: bool, optional
If True, it interprets the values passed are from a circular variable measured in radians and a circular KDE is used. Only valid for 1D KDE. Defaults to False. Only works if
kind == kde.- bins: integer or sequence or ‘auto’, optional
Controls the number of bins,accepts the same keywords
matplotlib.pyplot.hist()does. Only works ifkind == hist. If None (default) it will useautofor continuous variables andrange(xmin, xmax + 1)for discrete variables.- labeller
labellerinstance, optional Class providing the method
make_label_vertto generate the labels in the plot titles. Read the Label guide for more details and usage examples.- ax: numpy array-like of matplotlib axes or bokeh figures, optional
A 2D array of locations into which to plot the densities. If not supplied, Arviz will create its own array of plot areas (and return it).
- backend: str, optional
Select plotting backend {“matplotlib”,”bokeh”}. Default “matplotlib”.
- backend_kwargs: bool, optional
These are kwargs specific to the backend being used, passed to
matplotlib.pyplot.subplots()orbokeh.plotting.figure()- show: bool, optional
Call backend show function.
- **kwargs
Passed as-is to
matplotlib.pyplot.hist()ormatplotlib.pyplot.plot()function depending on the value ofkind.
- Returns:
- axes:
matplotlibaxesorbokehfigures
- axes:
See also
plot_distPlot distribution as histogram or kernel density estimates.
plot_densityGenerate KDE plots for continuous variables and histograms for discrete ones.
plot_forestForest plot to compare HDI intervals from a number of distributions.
Examples
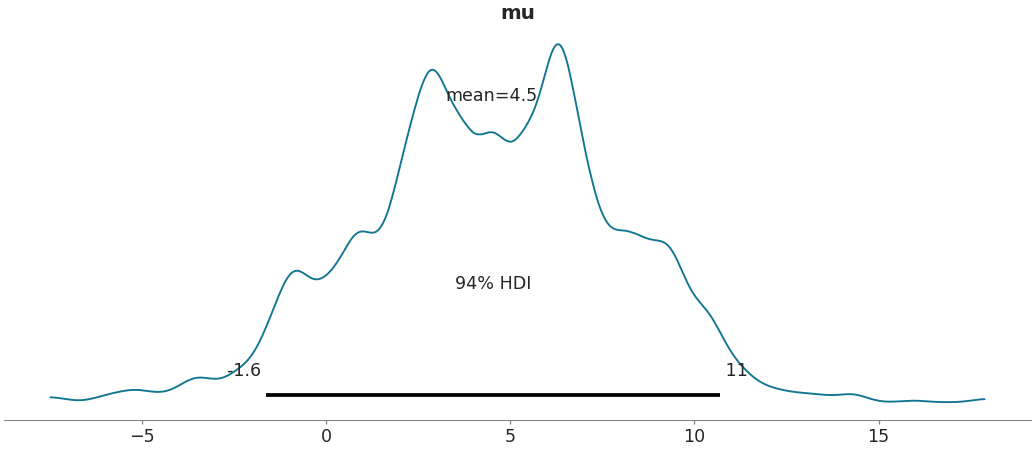
Show a default kernel density plot following style of John Kruschke
>>> import arviz as az >>> data = az.load_arviz_data('centered_eight') >>> az.plot_posterior(data)
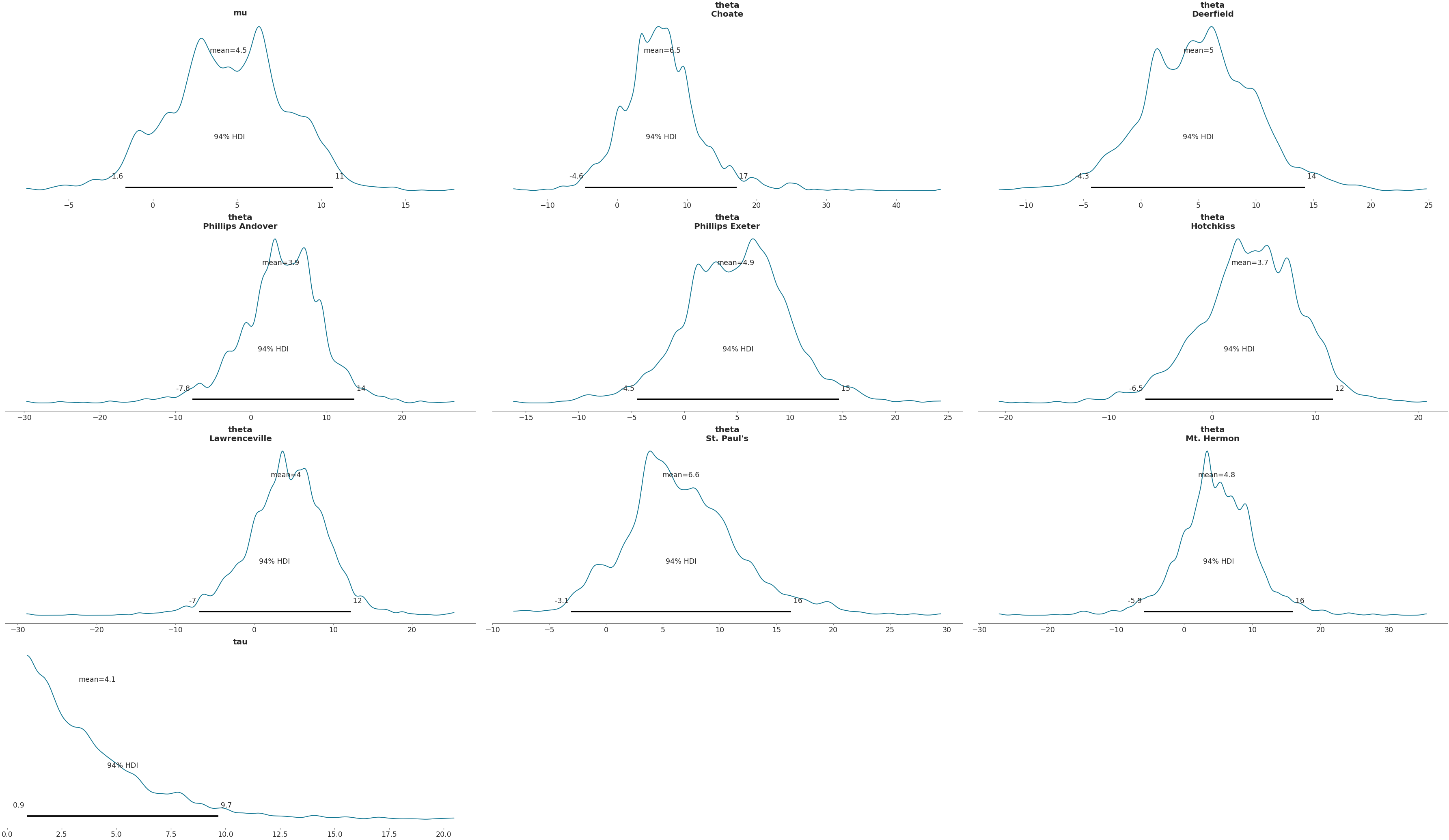
Plot subset variables by specifying variable name exactly
>>> az.plot_posterior(data, var_names=['mu'])
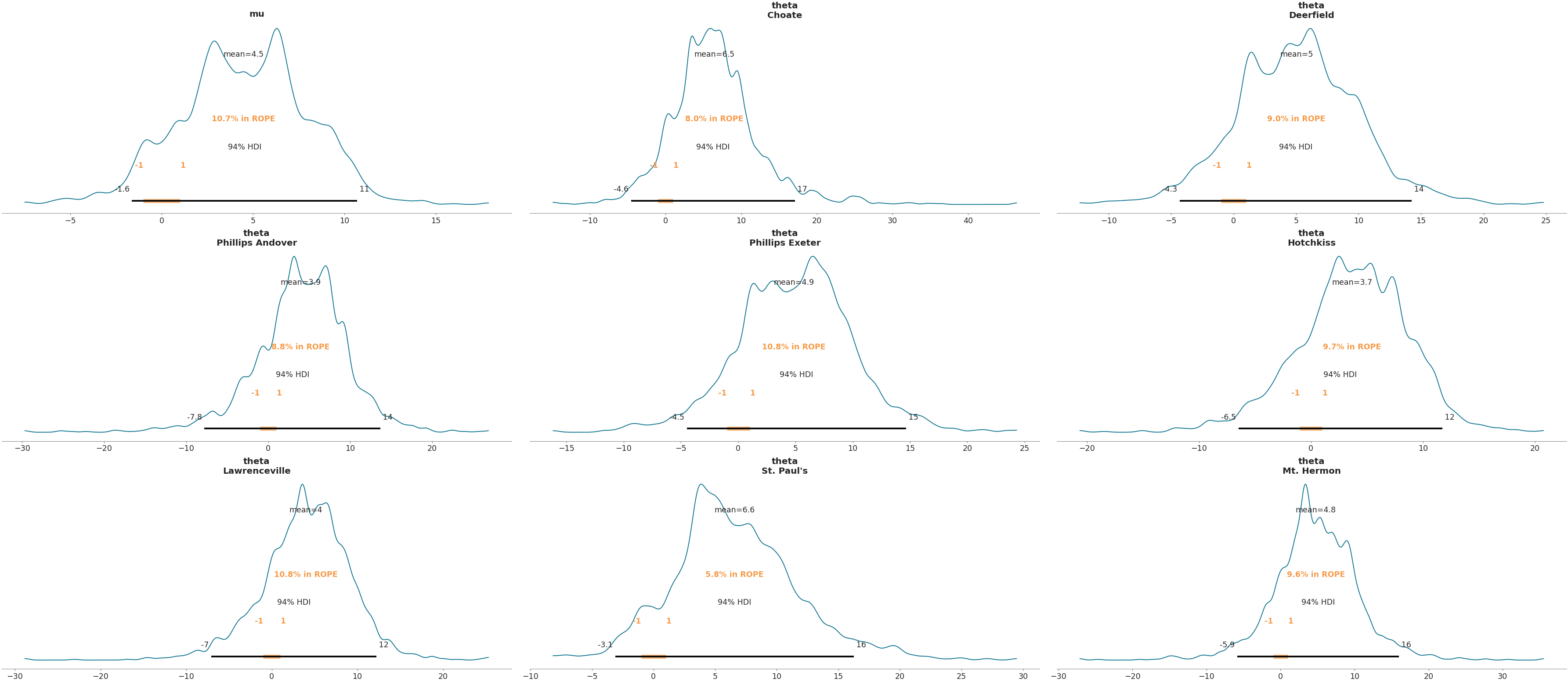
Plot Region of Practical Equivalence (rope) and select variables with regular expressions
>>> az.plot_posterior(data, var_names=['mu', '^the'], filter_vars="regex", rope=(-1, 1))
Plot Region of Practical Equivalence for selected distributions
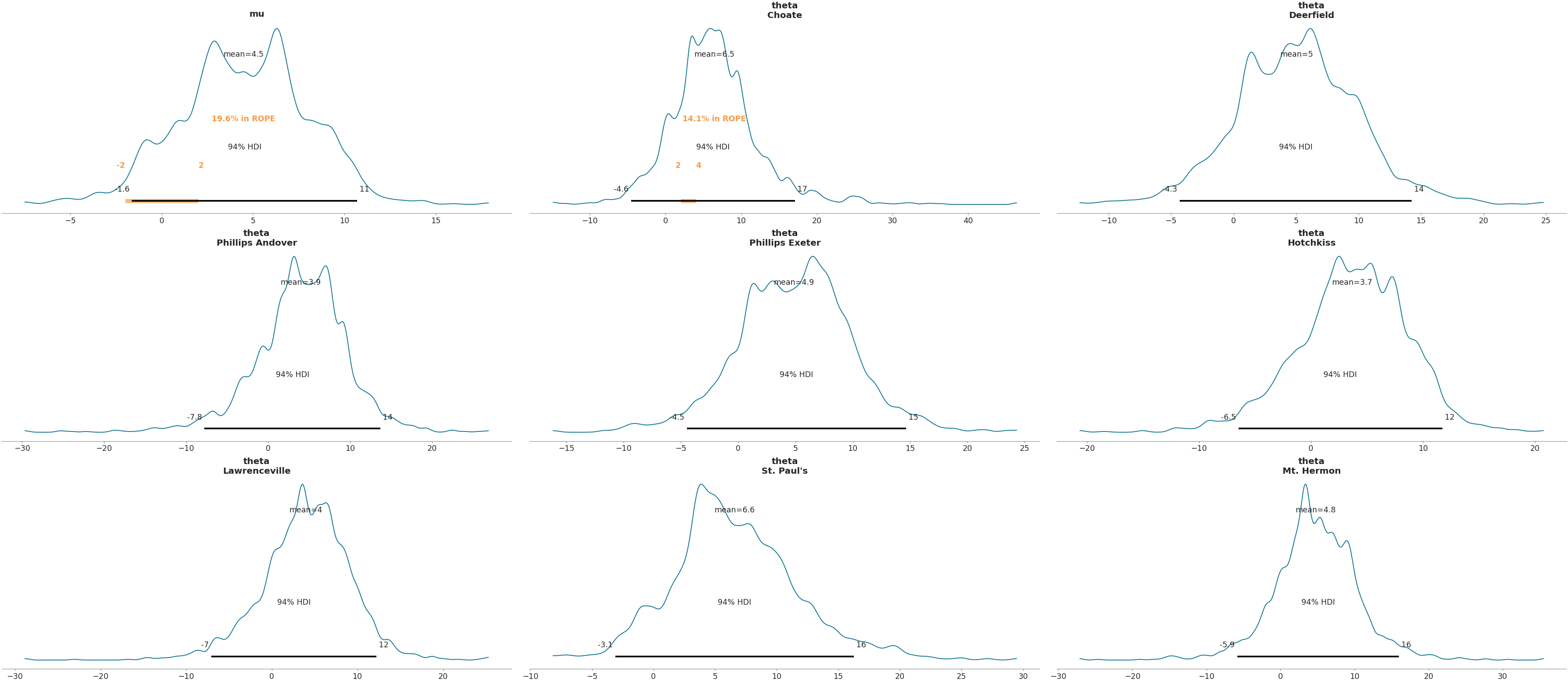
>>> rope = {'mu': [{'rope': (-2, 2)}], 'theta': [{'school': 'Choate', 'rope': (2, 4)}]} >>> az.plot_posterior(data, var_names=['mu', 'theta'], rope=rope)
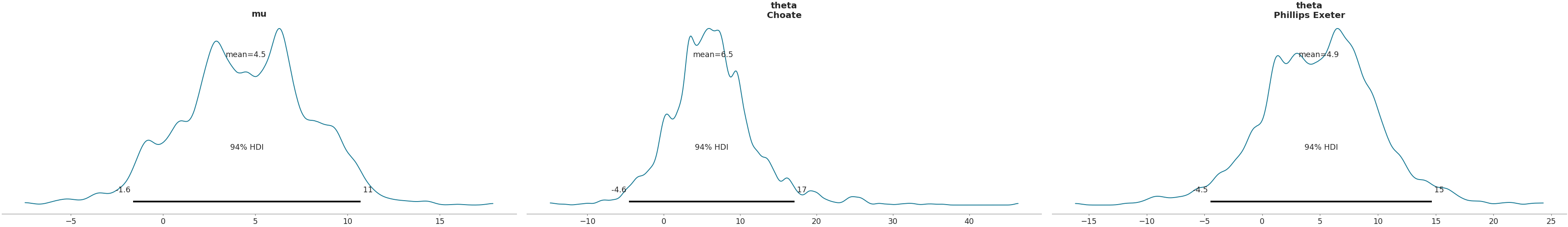
Using
coordsargument to plot only a subset of data>>> coords = {"school": ["Choate","Phillips Exeter"]} >>> az.plot_posterior(data, var_names=["mu", "theta"], coords=coords)
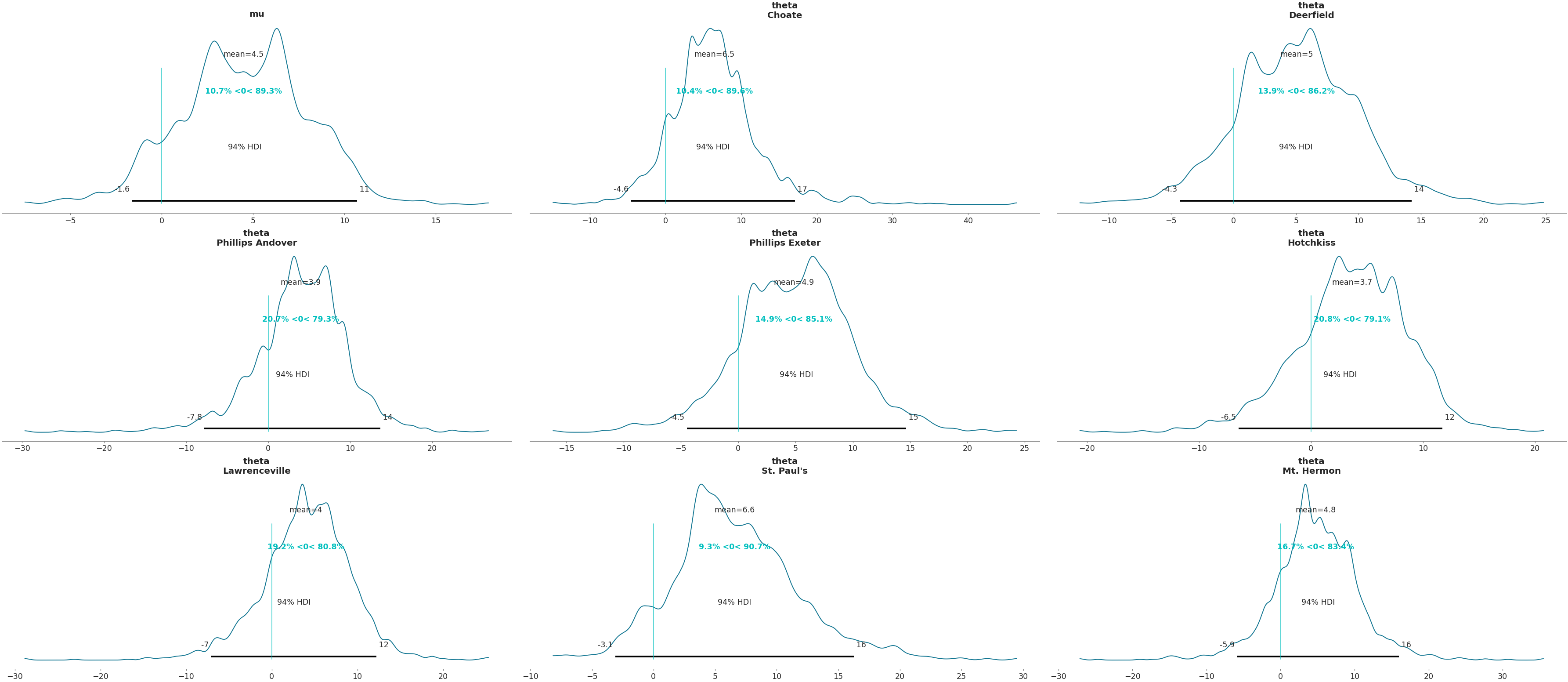
Add reference lines
>>> az.plot_posterior(data, var_names=['mu', 'theta'], ref_val=0)
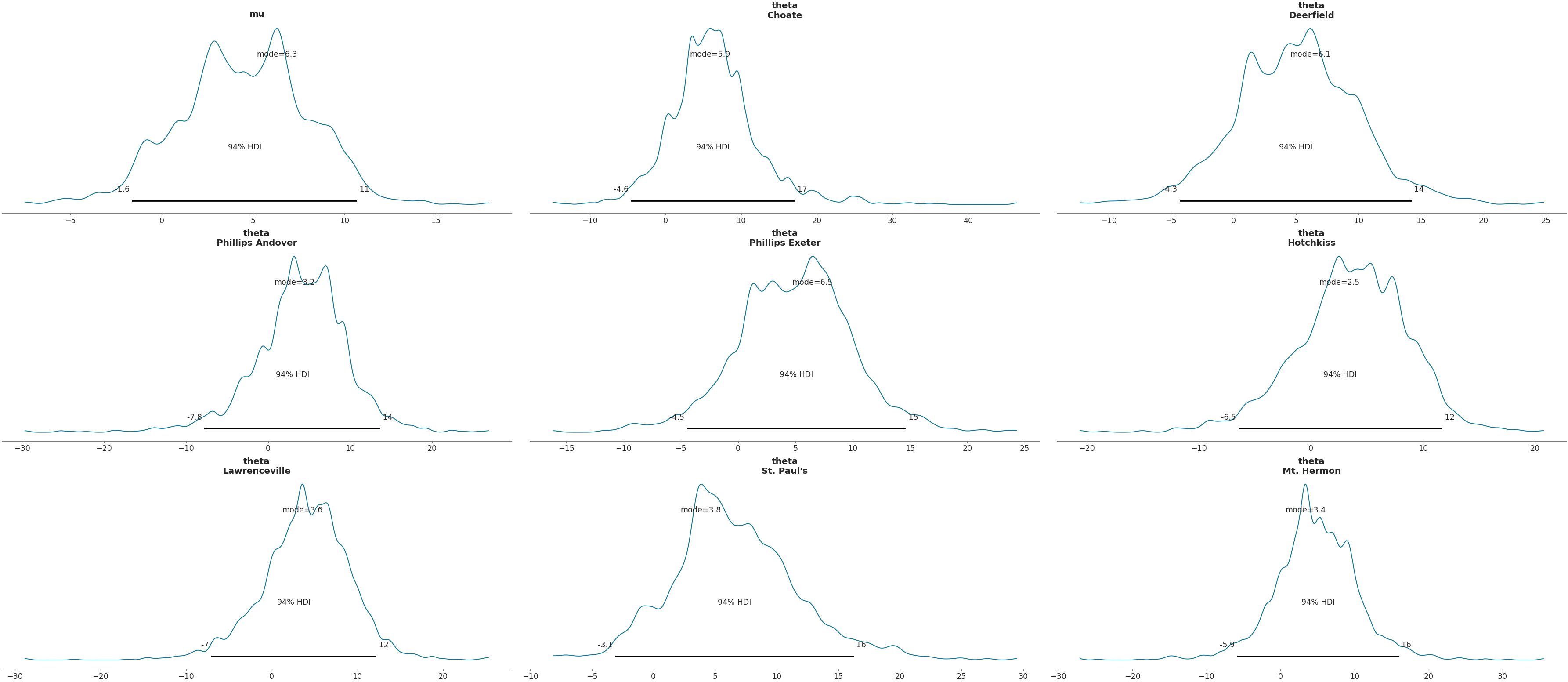
Show point estimate of distribution
>>> az.plot_posterior(data, var_names=['mu', 'theta'], point_estimate='mode')
Show reference values using variable names and coordinates
>>> az.plot_posterior(data, ref_val= {"theta": [{"school": "Deerfield", "ref_val": 4}, ... {"school": "Choate", "ref_val": 3}]})
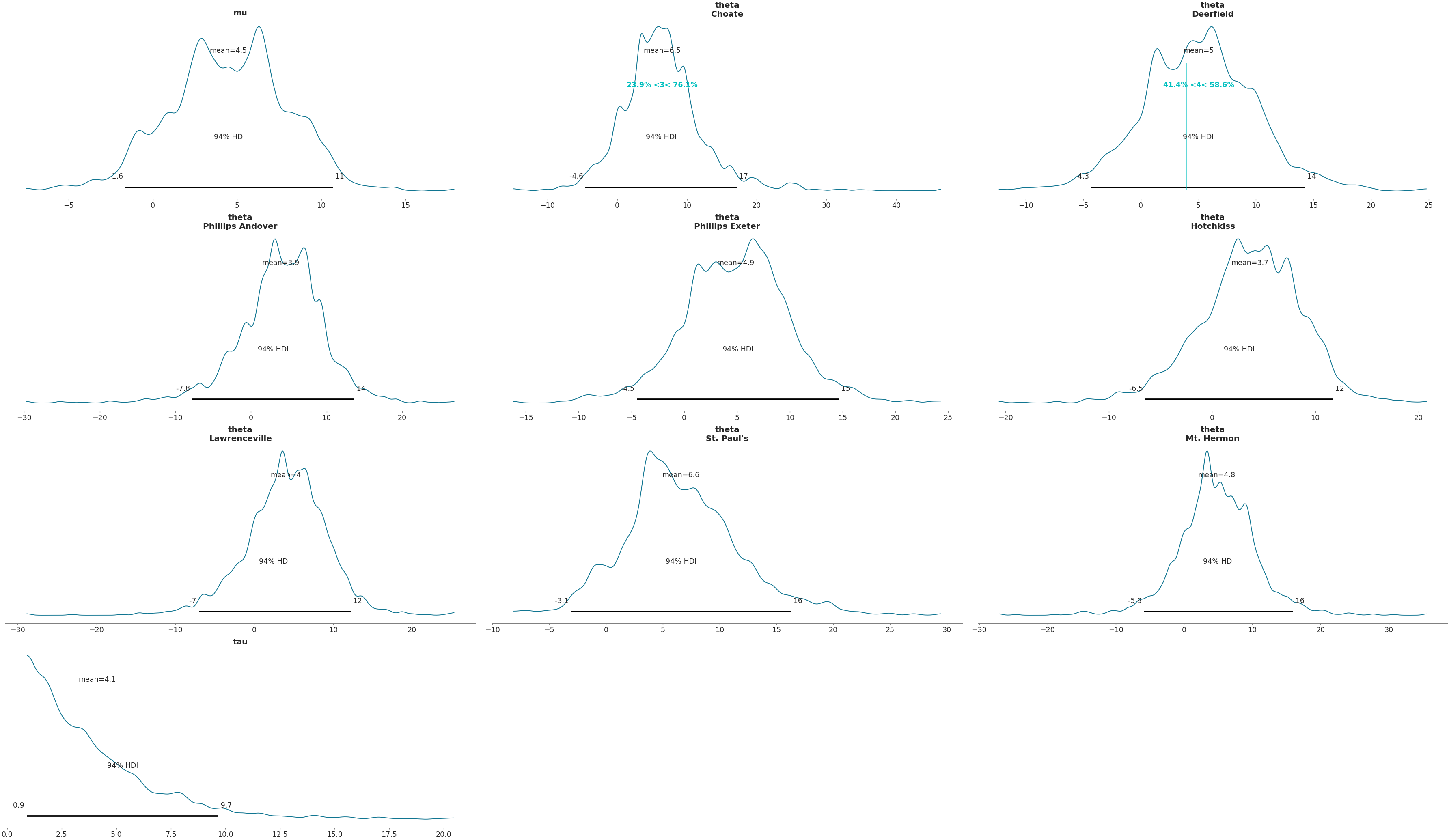
Show reference values using a list
>>> az.plot_posterior(data, ref_val=[1] + [5] * 8 + [1])
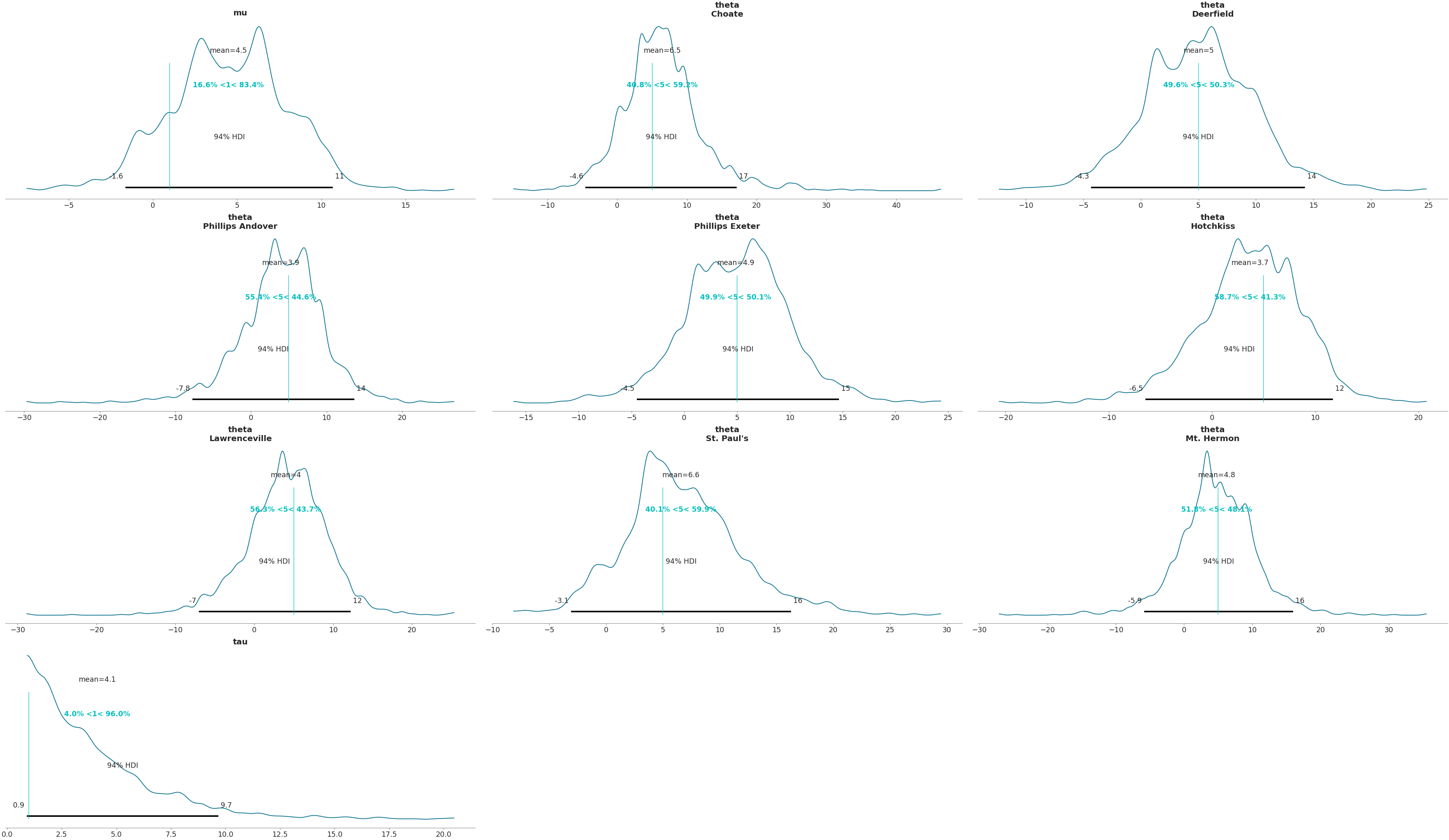
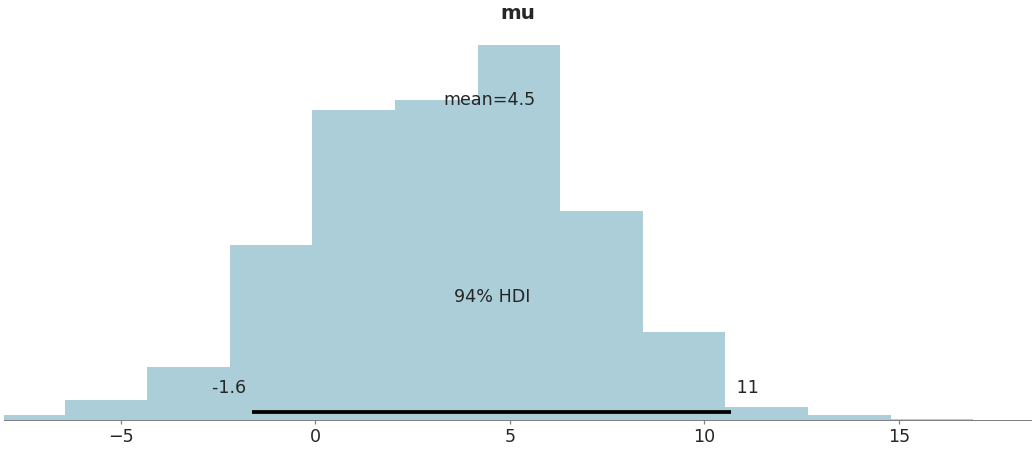
Plot posterior as a histogram
>>> az.plot_posterior(data, var_names=['mu'], kind='hist')
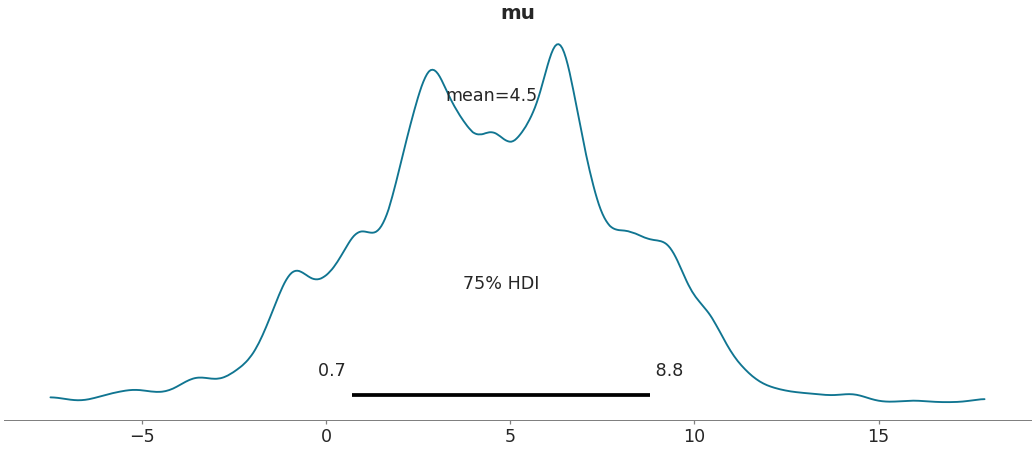
Change size of highest density interval
>>> az.plot_posterior(data, var_names=['mu'], hdi_prob=.75)