arviz.plot_bpv#
- arviz.plot_bpv(data, kind='u_value', t_stat='median', bpv=True, plot_mean=True, reference='analytical', mse=False, n_ref=100, hdi_prob=0.94, color='C0', grid=None, figsize=None, textsize=None, labeller=None, data_pairs=None, var_names=None, filter_vars=None, coords=None, flatten=None, flatten_pp=None, ax=None, backend=None, plot_ref_kwargs=None, backend_kwargs=None, group='posterior', show=None)[source]#
Plot Bayesian p-value for observed data and Posterior/Prior predictive.
- Parameters:
- data
InferenceData arviz.InferenceDataobject containing the observed and posterior/prior predictive data.- kind{“u_value”, “p_value”, “t_stat”}, default “u_value”
Specify the kind of plot:
The
kind="p_value"computes \(p := p(y* \leq y | y)\). This is the probability of the data y being larger or equal than the predicted data y*. The ideal value is 0.5 (half the predictions below and half above the data).The
kind="u_value"argument computes \(p_i := p(y_i* \leq y_i | y)\). i.e. like a p_value but per observation \(y_i\). This is also known as marginal p_value. The ideal distribution is uniform. This is similar to the LOO-PIT calculation/plot, the difference is than in LOO-pit plot we compute \(pi = p(y_i* r \leq y_i | y_{-i} )\), where \(y_{-i}\), is all other data except \(y_i\).The
kind="t_stat"argument computes \(:= p(T(y)* \leq T(y) | y)\) where T is any test statistic. Seet_statargument below for details of available options.
- t_stat
str,float, orcallable(), default “median” Test statistics to compute from the observations and predictive distributions. Allowed strings are “mean”, “median” or “std”. Alternative a quantile can be passed as a float (or str) in the interval (0, 1). Finally a user defined function is also acepted, see examples section for details.
- bpvbool, default
True If True add the Bayesian p_value to the legend when
kind = t_stat.- plot_meanbool, default
True Whether or not to plot the mean test statistic.
- reference{“analytical”, “samples”,
None}, default “analytical” How to compute the distributions used as reference for
kind=u_valuesorkind=p_values. UseNoneto not plot any reference.- msebool, default
False Show scaled mean square error between uniform distribution and marginal p_value distribution.
- n_ref
int, default 100 Number of reference distributions to sample when
reference=samples.- hdi_prob
float, optional Probability for the highest density interval for the analytical reference distribution when
kind=u_values. Should be in the interval (0, 1]. Defaults to the rcParamstats.hdi_prob. See this section for usage examples.- color
str, optional Matplotlib color
- grid
tuple, optional Number of rows and columns. By default, the rows and columns are automatically inferred. See this section for usage examples.
- figsize(
float,float), optional Figure size. If None it will be defined automatically.
- textsize
float, optional Text size scaling factor for labels, titles and lines. If None it will be autoscaled based on
figsize.- data_pairs
dict, optional Dictionary containing relations between observed data and posterior/prior predictive data. Dictionary structure:
key = data var_name
value = posterior/prior predictive var_name
For example,
data_pairs = {'y' : 'y_hat'}If None, it will assume that the observed data and the posterior/prior predictive data have the same variable name.- LabellerLabeller, optional
Class providing the method
make_pp_labelto generate the labels in the plot titles. Read the Label guide for more details and usage examples.- var_names
listofstr, optional Variables to be plotted. If
Noneall variable are plotted. Prefix the variables by~when you want to exclude them from the plot. See the this section for usage examples. See this section for usage examples.- filter_vars{
None, “like”, “regex”}, defaultNone If
None(default), interpretvar_namesas the real variables names. If “like”, interpretvar_namesas substrings of the real variables names. If “regex”, interpretvar_namesas regular expressions on the real variables names. See this section for usage examples.- coords
dict, optional Dictionary mapping dimensions to selected coordinates to be plotted. Dimensions without a mapping specified will include all coordinates for that dimension. Defaults to including all coordinates for all dimensions if None. See this section for usage examples.
- flatten
list, optional List of dimensions to flatten in observed_data. Only flattens across the coordinates specified in the coords argument. Defaults to flattening all of the dimensions.
- flatten_pp
list, optional List of dimensions to flatten in posterior_predictive/prior_predictive. Only flattens across the coordinates specified in the coords argument. Defaults to flattening all of the dimensions. Dimensions should match flatten excluding dimensions for data_pairs parameters. If
flattenis defined andflatten_ppis None, thenflatten_pp=flatten.- legendbool, default
True Add legend to figure.
- ax2D array_like of
matplotlib AxesorBokeh Figure, optional A 2D array of locations into which to plot the densities. If not supplied, ArviZ will create its own array of plot areas (and return it).
- backend
str, optional Select plotting backend {“matplotlib”, “bokeh”}. Default “matplotlib”.
- plot_ref_kwargs
dict, optional Extra keyword arguments to control how reference is represented. Passed to
matplotlib.axes.Axes.plot()ormatplotlib.axes.Axes.axhspan()(whenkind=u_valueandreference=analytical).- backend_kwargsbool, optional
These are kwargs specific to the backend being used, passed to
matplotlib.pyplot.subplots()orbokeh.plotting.figure. For additional documentation check the plotting method of the backend.- group{“posterior”, “prior”}, default “posterior”
Specifies which InferenceData group should be plotted. If “posterior”, then the values in
posterior_predictivegroup are compared to the ones inobserved_data, if “prior” then the same comparison happens, but with the values inprior_predictivegroup.- showbool, optional
Call backend show function.
- data
- Returns:
- axes2D
ndarrayofmatplotlib AxesorBokeh Figure
- axes2D
See also
plot_ppcPlot for posterior/prior predictive checks.
plot_loo_pitPlot Leave-One-Out probability integral transformation (PIT) predictive checks.
plot_dist_comparisonPlot to compare fitted and unfitted distributions.
Notes
Discrete data is smoothed before computing either p-values or u-values using the function
smooth_data()References
Gelman et al. (2013) see http://www.stat.columbia.edu/~gelman/book/ pages 151-153 for details
Examples
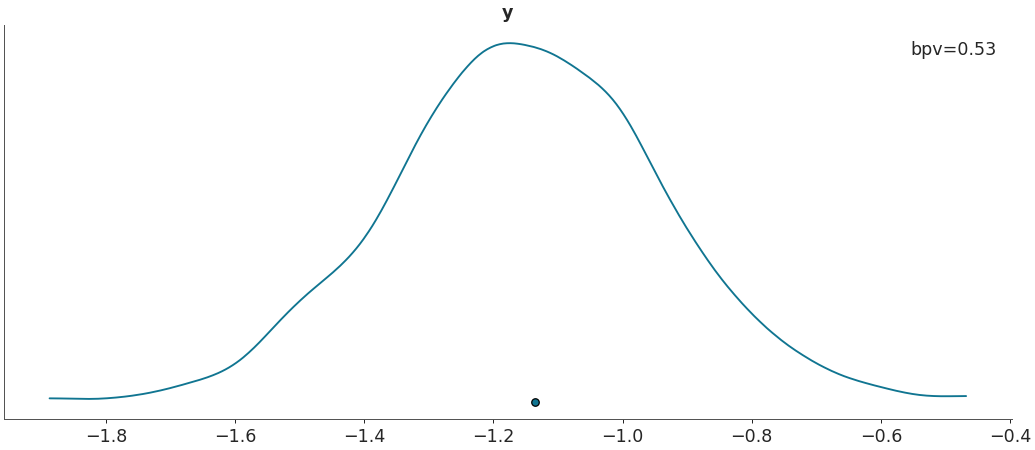
Plot Bayesian p_values.
>>> import arviz as az >>> data = az.load_arviz_data("regression1d") >>> az.plot_bpv(data, kind="p_value")
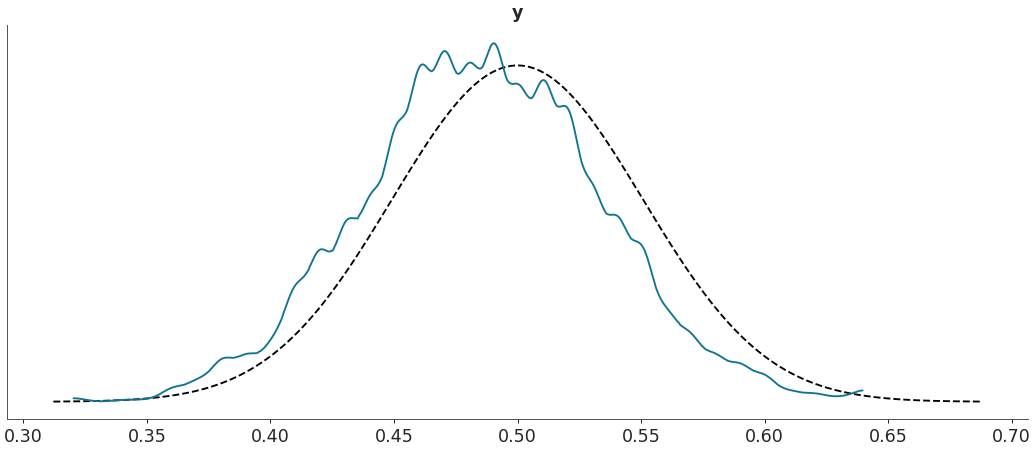
Plot custom test statistic comparison.
>>> import arviz as az >>> data = az.load_arviz_data("regression1d") >>> az.plot_bpv(data, kind="t_stat", t_stat=lambda x:np.percentile(x, q=50, axis=-1))