arviz.kde#
- arviz.kde(x, circular=False, **kwargs)[source]#
One dimensional density estimation.
It is a wrapper around
kde_linear()andkde_circular().- Parameters:
- Returns:
- grid
numpy.ndarray Gridded numpy array for the x values.
- pdf
numpy.ndarray Numpy array for the density estimates.
- bw
float The estimated bandwidth. Only returned if requested.
- grid
See also
plot_kdeCompute and plot a kernel density estimate.
Examples
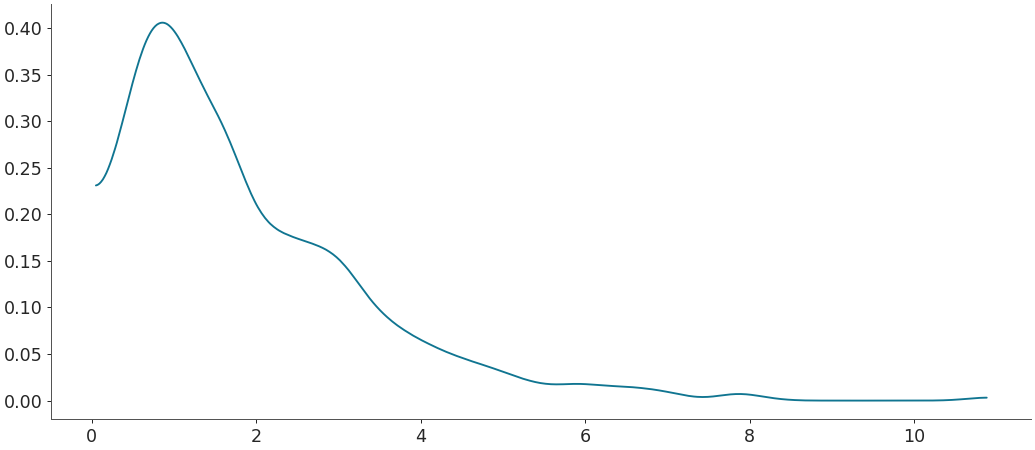
Default density estimation for linear data
>>> import numpy as np >>> import matplotlib.pyplot as plt >>> from arviz import kde >>> >>> rng = np.random.default_rng(49) >>> rvs = rng.gamma(shape=1.8, size=1000) >>> grid, pdf = kde(rvs) >>> plt.plot(grid, pdf)
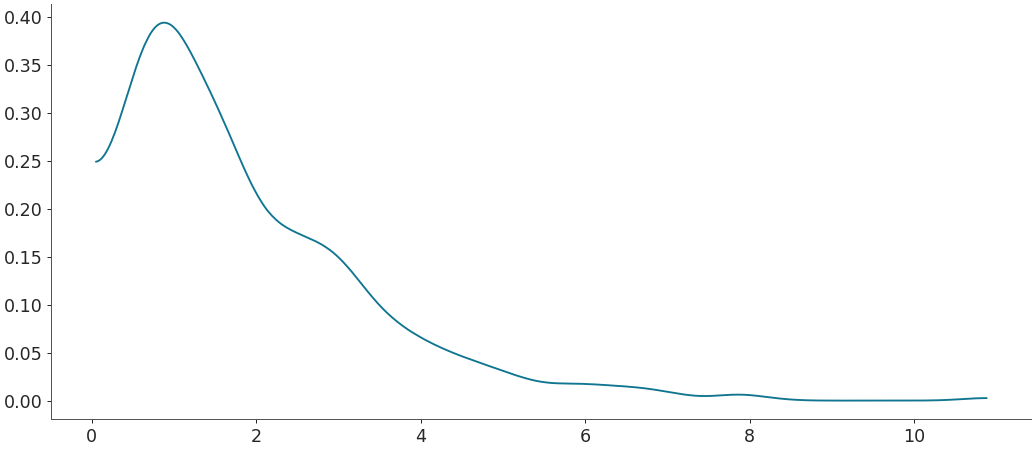
Density estimation for linear data with Silverman’s rule bandwidth
>>> grid, pdf = kde(rvs, bw="silverman") >>> plt.plot(grid, pdf)
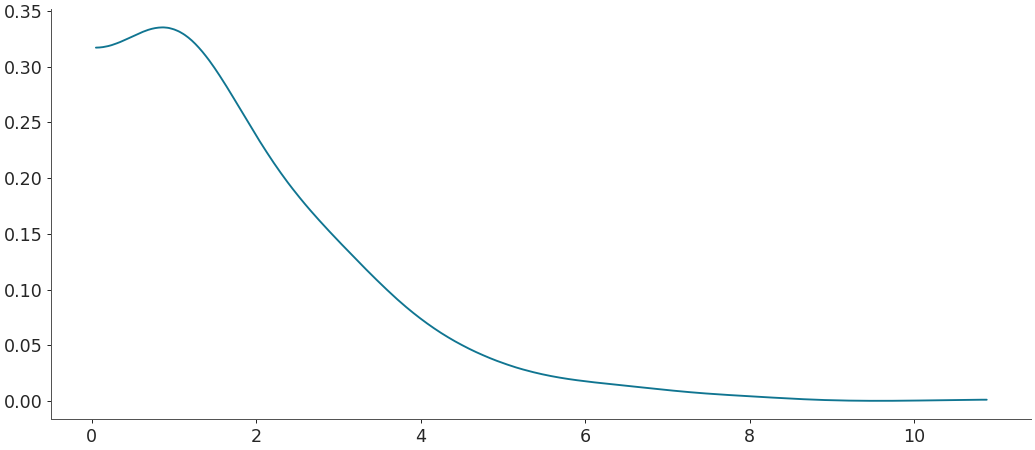
Density estimation for linear data with scaled bandwidth
>>> # bw_fct > 1 means more smoothness. >>> grid, pdf = kde(rvs, bw_fct=2.5) >>> plt.plot(grid, pdf)
Default density estimation for linear data with extended limits
>>> grid, pdf = kde(rvs, bound_correction=False, extend=True, extend_fct=0.5) >>> plt.plot(grid, pdf)
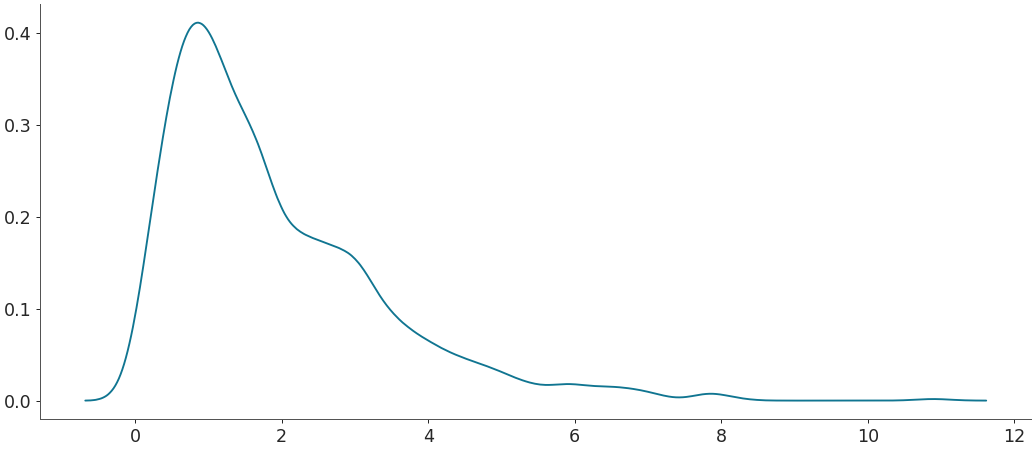
Default density estimation for linear data with custom limits
>>> # It accepts tuples and lists of length 2. >>> grid, pdf = kde(rvs, bound_correction=False, custom_lims=(0, 11)) >>> plt.plot(grid, pdf)
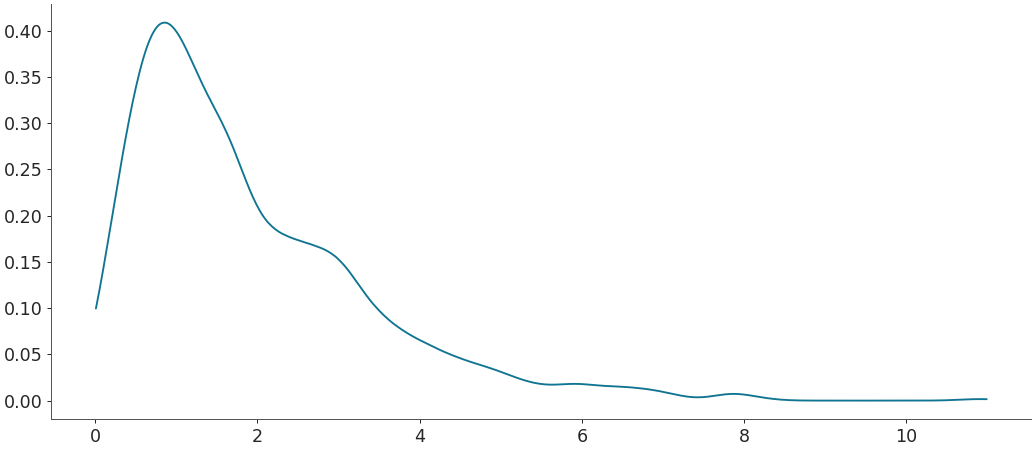
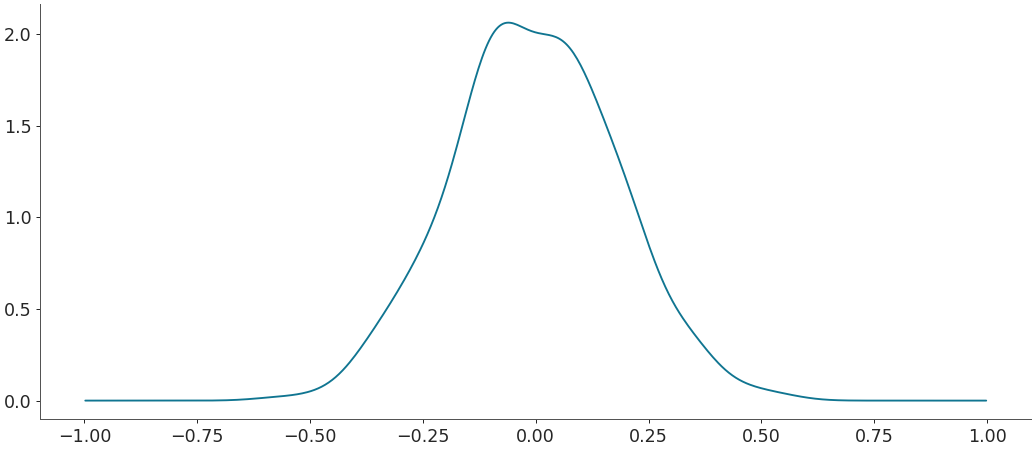
Default density estimation for circular data
>>> rvs = np.random.vonmises(mu=np.pi, kappa=1, size=500) >>> grid, pdf = kde(rvs, circular=True) >>> plt.plot(grid, pdf)
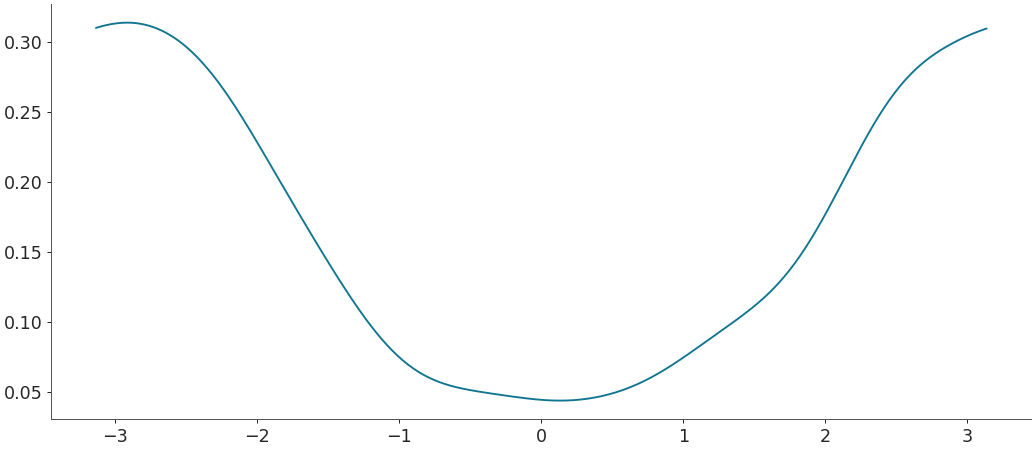
Density estimation for circular data with scaled bandwidth
>>> rvs = np.random.vonmises(mu=np.pi, kappa=1, size=500) >>> # bw_fct > 1 means less smoothness. >>> grid, pdf = kde(rvs, circular=True, bw_fct=3) >>> plt.plot(grid, pdf)
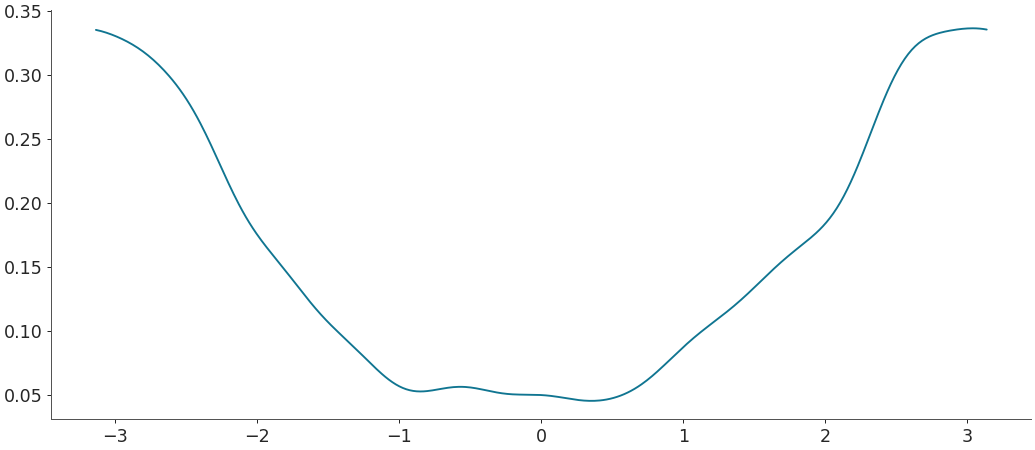
Density estimation for circular data with custom limits
>>> # This is still experimental, does not always work. >>> rvs = np.random.vonmises(mu=0, kappa=30, size=500) >>> grid, pdf = kde(rvs, circular=True, custom_lims=(-1, 1)) >>> plt.plot(grid, pdf)