arviz.plot_hdi#
- arviz.plot_hdi(x, y=None, hdi_prob=None, hdi_data=None, color='C1', circular=False, smooth=True, smooth_kwargs=None, figsize=None, fill_kwargs=None, plot_kwargs=None, hdi_kwargs=None, ax=None, backend=None, backend_kwargs=None, show=None)[source]#
Plot HDI intervals for regression data.
- Parameters:
- xarray_like
Values to plot.
- yarray_like, optional
Values from which to compute the HDI. Assumed shape
(chain, draw, \*shape). Only optional ifhdi_datais present.- hdi_dataarray_like, optional
Precomputed HDI values to use. Assumed shape is
(*x.shape, 2).- hdi_prob
float, optional Probability for the highest density interval. Defaults to
stats.hdi_probrcParam. See this section for usage examples.- color
str, default “C1” Color used for the limits of the HDI and fill. Should be a valid matplotlib color.
- circularbool, default
False Whether to compute the HDI taking into account
xis a circular variable (in the range [-np.pi, np.pi]) or not. Defaults to False (i.e non-circular variables).- smoothbool, default
True If True the result will be smoothed by first computing a linear interpolation of the data over a regular grid and then applying the Savitzky-Golay filter to the interpolated data.
- smooth_kwargs
dict, optional Additional keywords modifying the Savitzky-Golay filter. See
scipy.signal.savgol_filter()for details.- figsize(
float,float), optional Figure size. If
None, it will be defined automatically.- fill_kwargs
dict, optional Keywords passed to
matplotlib.axes.Axes.fill_between()(usefill_kwargs={'alpha': 0}to disable fill) or tobokeh.plotting.Figure.patch().- plot_kwargs
dict, optional HDI limits keyword arguments, passed to
matplotlib.axes.Axes.plot()orbokeh.plotting.Figure.patch().- hdi_kwargs
dict, optional Keyword arguments passed to
hdi(). Ignored ifhdi_datais present.- ax
axes, optional Matplotlib axes or bokeh figures.
- backend{“matplotlib”, “bokeh”}, default “matplotlib”
Select plotting backend.
- backend_kwargs
dict, optional These are kwargs specific to the backend being used, passed to
matplotlib.pyplot.subplots()orbokeh.plotting.figure. For additional documentation check the plotting method of the backend.- showbool, optional
Call backend show function.
- Returns:
- axes
matplotlibaxesorbokehfigures
- axes
See also
hdiCalculate highest density interval (HDI) of array for given probability.
Examples
Plot HDI interval of simulated random-walk data using
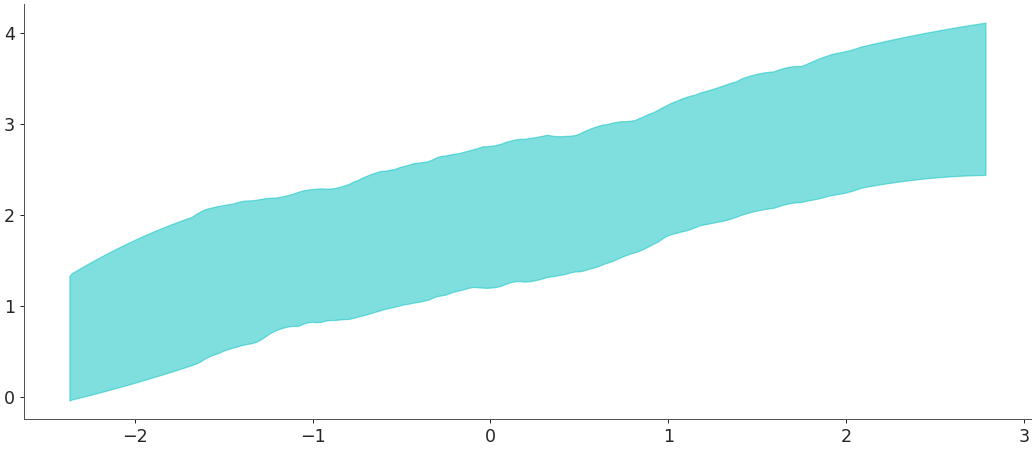
yargument:>>> import numpy as np >>> import arviz as az >>> # time-steps random walk >>> x_data =np.arange(0,100) >>> # Mean random walk >>> mu = np.zeros(100) >>> for i in x_data: mu[i] = mu[i-1] + np.random.normal(0, 1, 1) >>> # Simulated pp samples form the random walk time series >>> y_data = np.random.normal(2 + mu * 0.5, 0.5, size = (2, 50, 100)) >>> az.plot_hdi(x_data, y_data)
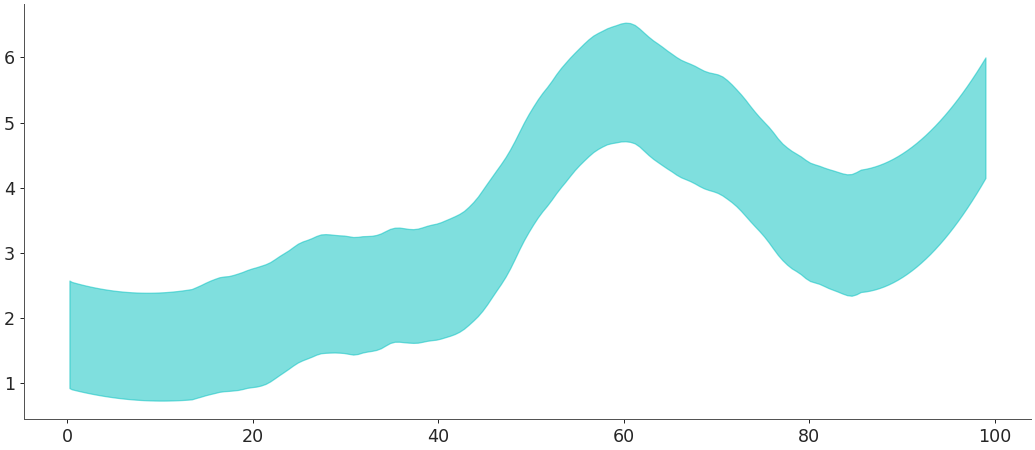
plot_hdican also be given precalculated values with the argumenthdi_data. This example shows how to usehdi()to precalculate the values and pass these values toplot_hdi. Similarly to an example inhdiwe are using theinput_core_dimsargument ofwrap_xarray_ufunc()to manually define the dimensions over which to calculate the HDI.>>> hdi_data = az.hdi(y_data, input_core_dims=[["draw"]]) >>> ax = az.plot_hdi(x_data, hdi_data=hdi_data[0], color="r", fill_kwargs={"alpha": .2}) >>> az.plot_hdi(x_data, hdi_data=hdi_data[1], color="k", ax=ax, fill_kwargs={"alpha": .2})
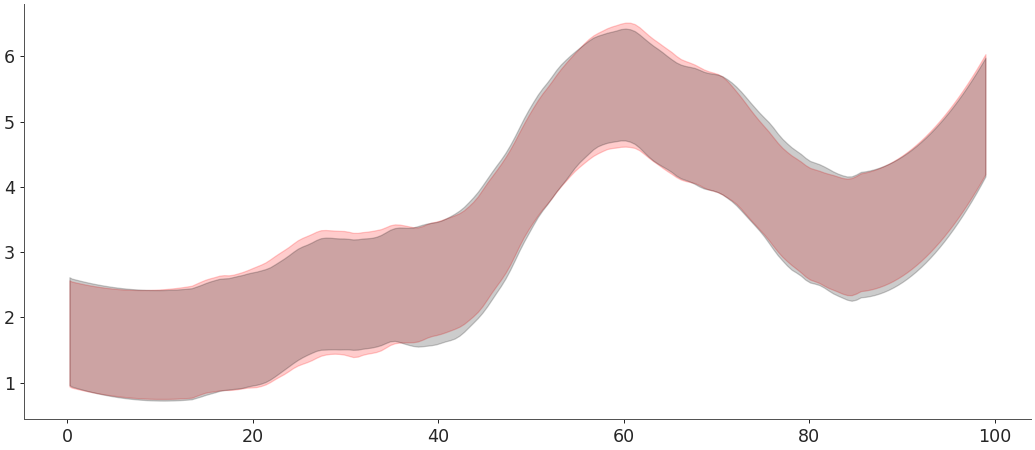
plot_hdican also be used with Inference Data objects. Here we use the posterior predictive to plot the HDI interval.>>> X = np.random.normal(0,1,100) >>> Y = np.random.normal(2 + X * 0.5, 0.5, size=(2,10,100)) >>> idata = az.from_dict(posterior={"y": Y}, constant_data={"x":X}) >>> x_data = idata.constant_data.x >>> y_data = idata.posterior.y >>> az.plot_hdi(x_data, y_data)