Dask for ArviZ#
Dask overview#
Dask is a big data processing library used for:
Parallelizing the computation of the workflow consisting of NumPy, pandas, xarray and scikit-learn frameworks.
Scaling the workflows up or down depending upon the hardware that is being used.
Most notably, it provides the support for working with larger-than-memory datasets. In this case, dask partitions the dataset into smaller chunks, then loads only a few chunks from the disk, and once the necessary processing is completed, it throws away the intermediate values. This way, the computations are performed without exceeding the memory limit.
Check out these links if you’re unsure whether your workflow can benefit from using Dask or not:
Excerpt from “Dask Array Best Practices” doc.
If your data fits comfortably in RAM and you are not performance bound, then using
NumPymight be the right choice. Dask adds another layer of complexity which may get in the way.If you are just looking for speedups rather than scalability then you may want to consider a project like
Numba.
Caution
Dask is an optional dependency inside ArviZ, which is still being actively developed. Currently, few functions belonging to diagnostics and stats module can utilize Dask’s capabilities.
# optional imports
from dask.distributed import Client
from dask.diagnostics import ResourceProfiler
from bokeh.resources import INLINE
import bokeh.io
bokeh.io.output_notebook(INLINE)
%reload_ext memory_profiler
Note
ResourceProfiler() and Client are optional. They are only used for the visualizing and profiling the dask enabled methods. ArviZ-Dask integration can be used without using these objects.
client = Client(threads_per_worker=4, n_workers=1, memory_limit="1.2GB")
client
Client
Client-3a9af271-4541-11ec-b824-5820b17a12fa
| Connection method: Cluster object | Cluster type: distributed.LocalCluster |
| Dashboard: http://127.0.0.1:8787/status |
Cluster Info
LocalCluster
58ea9317
| Dashboard: http://127.0.0.1:8787/status | Workers: 1 |
| Total threads: 4 | Total memory: 1.12 GiB |
| Status: running | Using processes: True |
Scheduler Info
Scheduler
Scheduler-b3987f5c-3da8-4564-9fb5-76fb51a47a9e
| Comm: tcp://127.0.0.1:35215 | Workers: 1 |
| Dashboard: http://127.0.0.1:8787/status | Total threads: 4 |
| Started: Just now | Total memory: 1.12 GiB |
Workers
Worker: 0
| Comm: tcp://127.0.0.1:36217 | Total threads: 4 |
| Dashboard: http://127.0.0.1:37745/status | Memory: 1.12 GiB |
| Nanny: tcp://127.0.0.1:41547 | |
| Local directory: /home/oriol/Public/arviz/doc/source/user_guide/dask-worker-space/worker-zyz5nqyr | |
Variance example#
array_size = 250_000_000
Calculating variance using Numpy
%%memit
data = np.random.randn(array_size)
np.var(data, ddof=1)
del data
peak memory: 4072.28 MiB, increment: 3815.28 MiB
Calculating variance using Dask arrays:
Divides the array into multiple chunks.
Objects are lazy in nature and are computed on-the-fly.
Builds a task graph of the entire computation and parallelizes the execution.
%memit data = dask.array.random.normal(size=array_size, chunks="auto")
data
peak memory: 258.30 MiB, increment: 0.28 MiB
|
var = dask.array.var(data, ddof=1)
var.visualize()
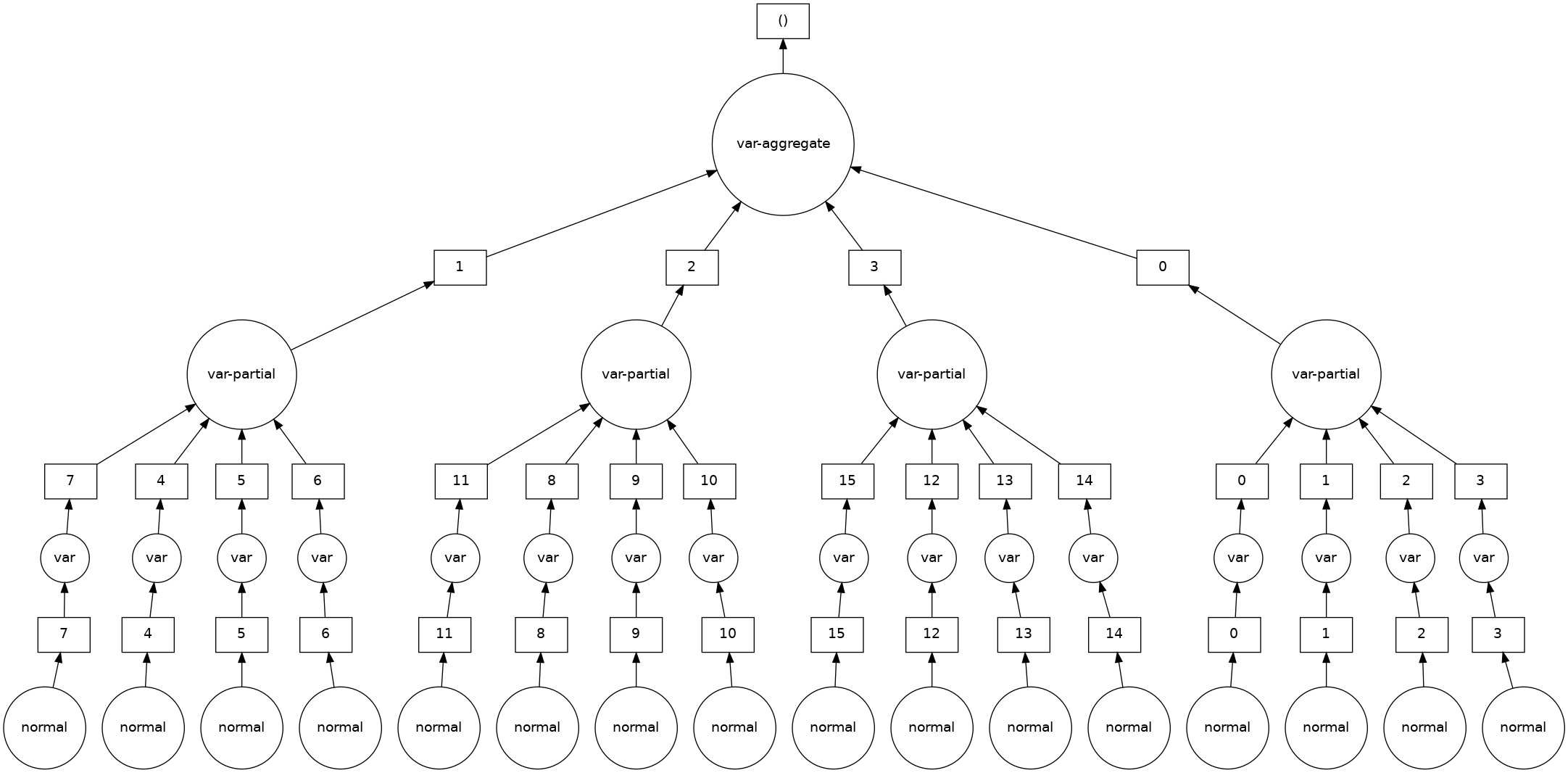
with ResourceProfiler(dt=0.25) as rprof:
var.compute()
rprof.visualize();
del data
Here, the NumPy version consumed around ~5GB memory but the Dask version was able to compute variance in under 1.2Gb memory (the limit set in the Client configuration above) which shows how beneficial Dask can be when dealing with large datasets.
ArviZ-Dask integration#
Creating Dask-backed InferenceData objects#
InferenceData is the central data format for ArviZ and there are several ways to generate this object (which you can look here.
However, as the ArviZ-Dask integraton is still a work in progress, to use InferenceData object with Dask-compatible methods we’ll have generate it in a different way. arviz.from_netcdf() has an experimental group_kwargs argument that can be used to read netCDF files directly with Dask.
We will progressively add more ways to generate Dask backed InferenceData and document them here. If you are interested in helping out, reach out on Gitter
From dictionary using dask.array#
We start creating a dask array with random samples, that we can then convert to InferenceData using arviz.from_dict(). ArviZ passes values and coord values as is to xarray, so by passing a dask array we’ll get a dask backed InferenceData automatically.
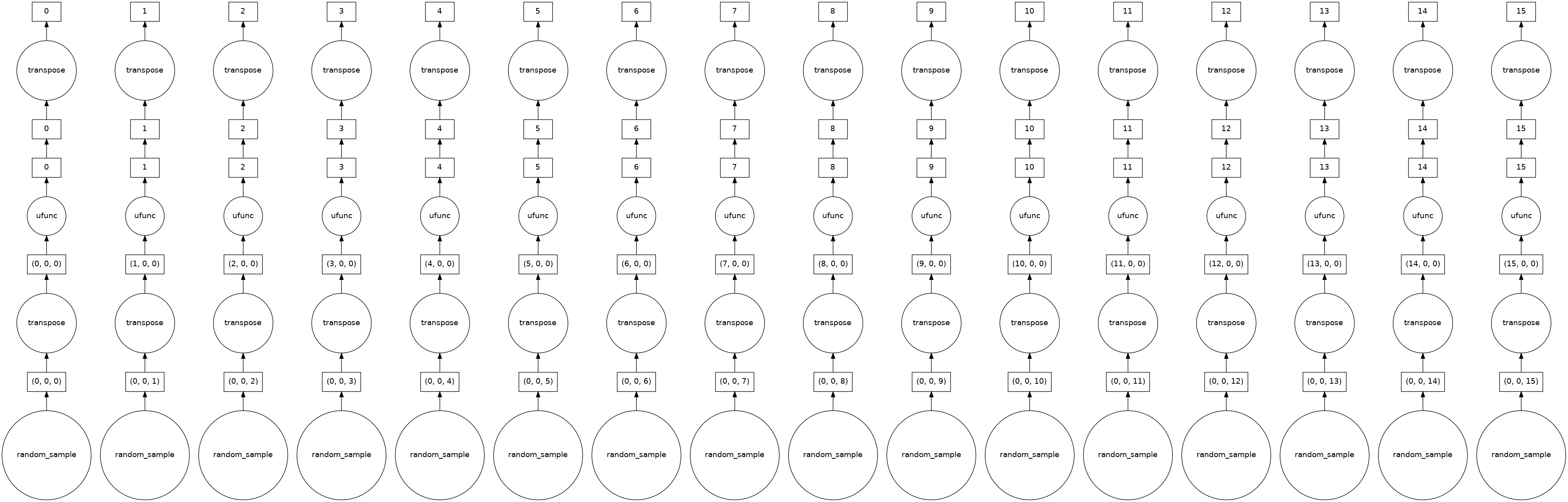
%memit daskdata = dask.array.random.random((10, 1000, 10000), chunks=(10, 1000, 625))
daskdata
peak memory: 260.43 MiB, increment: 0.07 MiB
|
daskdata.visualize() # Each chunk will follow lazy evaluation

Note
Setting up the right value of the chunks parameter is very important. Computation on Dask arrays with small chunks are slow because each operation on a chunk has some overhead. On the other side, if your chunks are too big, then it might not fit in the memory.
datadict = {"x": daskdata}
%memit idata_dask = az.from_dict(posterior=datadict, dims={"x": ["dim_1"]})
idata_dask
peak memory: 260.55 MiB, increment: 0.07 MiB
-
- chain: 10
- draw: 1000
- dim_1: 10000
- chain(chain)int640 1 2 3 4 5 6 7 8 9
array([0, 1, 2, 3, 4, 5, 6, 7, 8, 9])
- draw(draw)int640 1 2 3 4 5 ... 995 996 997 998 999
array([ 0, 1, 2, ..., 997, 998, 999])
- dim_1(dim_1)int640 1 2 3 4 ... 9996 9997 9998 9999
array([ 0, 1, 2, ..., 9997, 9998, 9999])
- x(chain, draw, dim_1)float64dask.array<chunksize=(10, 1000, 625), meta=np.ndarray>
Array Chunk Bytes 762.94 MiB 47.68 MiB Shape (10, 1000, 10000) (10, 1000, 625) Count 16 Tasks 16 Chunks Type float64 numpy.ndarray
- created_at :
- 2021-11-14T11:51:52.050185
- arviz_version :
- 0.11.4
<xarray.Dataset> Dimensions: (chain: 10, draw: 1000, dim_1: 10000) Coordinates: * chain (chain) int64 0 1 2 3 4 5 6 7 8 9 * draw (draw) int64 0 1 2 3 4 5 6 7 8 ... 992 993 994 995 996 997 998 999 * dim_1 (dim_1) int64 0 1 2 3 4 5 6 ... 9993 9994 9995 9996 9997 9998 9999 Data variables: x (chain, draw, dim_1) float64 dask.array<chunksize=(10, 1000, 625), meta=np.ndarray> Attributes: created_at: 2021-11-14T11:51:52.050185 arviz_version: 0.11.4xarray.Dataset
Executing ArviZ functions with Dask#
arviz.Dask provides the functionality of disabling/re-enabling Dask. This is an ArviZ specific class that therefore works only with ArviZ functions that support computation via Dask.
We can also use it to set default arguments which are then taken by the Dask supporting functions and passed to xarray.apply_ufunc().
For comparison lets first create an InferenceData object using numpy array
%memit npdata = np.random.rand(10, 1000, 10000)
datadict = {"x": npdata}
idata_numpy = az.from_dict(posterior=datadict, dims={"x": ["dim_1"]})
idata_numpy
peak memory: 1023.65 MiB, increment: 762.97 MiB
-
- chain: 10
- draw: 1000
- dim_1: 10000
- chain(chain)int640 1 2 3 4 5 6 7 8 9
array([0, 1, 2, 3, 4, 5, 6, 7, 8, 9])
- draw(draw)int640 1 2 3 4 5 ... 995 996 997 998 999
array([ 0, 1, 2, ..., 997, 998, 999])
- dim_1(dim_1)int640 1 2 3 4 ... 9996 9997 9998 9999
array([ 0, 1, 2, ..., 9997, 9998, 9999])
- x(chain, draw, dim_1)float640.3969 0.9378 ... 0.8059 0.6392
array([[[3.96893013e-01, 9.37832731e-01, 3.49005231e-01, ..., 1.36533029e-01, 8.52493391e-01, 3.11825216e-01], [2.24438848e-01, 5.46843569e-01, 2.23949351e-01, ..., 8.19941477e-01, 8.64443797e-02, 1.92593403e-01], [3.27913875e-01, 7.92223340e-01, 8.45441266e-02, ..., 5.21305713e-01, 4.10688001e-01, 2.04838186e-01], ..., [5.83650829e-01, 7.22516288e-01, 1.99610883e-01, ..., 1.31725612e-01, 5.43205032e-01, 4.61174897e-01], [4.13712821e-01, 5.41706904e-01, 2.99458008e-01, ..., 8.80293621e-01, 9.73619510e-01, 5.86407345e-01], [6.12662482e-01, 8.02328683e-01, 9.69486169e-01, ..., 2.98126426e-01, 2.79703190e-02, 9.26846418e-04]], [[2.05484112e-01, 6.13739789e-01, 6.21671265e-01, ..., 7.92393616e-01, 6.80348978e-01, 7.38175852e-01], [6.37058780e-01, 7.88523683e-01, 2.75056811e-02, ..., 7.70084758e-01, 6.15943940e-02, 1.28364699e-01], [5.54628005e-01, 5.81644903e-01, 4.01931224e-01, ..., 5.30039904e-01, 2.35943604e-02, 6.49195282e-01], ... [3.34881965e-01, 8.01624356e-01, 7.55748346e-02, ..., 9.86300065e-01, 9.75312881e-01, 2.96055886e-01], [1.90045905e-01, 9.58935861e-01, 3.34628238e-01, ..., 2.40706967e-01, 8.63792822e-01, 9.38813997e-01], [7.83035223e-01, 6.98403375e-01, 5.63195463e-01, ..., 2.79195156e-02, 3.26374185e-01, 4.92441648e-01]], [[2.75316625e-01, 6.89981496e-01, 1.83490693e-01, ..., 7.75977562e-01, 7.87100253e-01, 1.56747858e-01], [3.68682670e-01, 2.75850782e-01, 2.69705447e-01, ..., 5.60678692e-01, 4.47949484e-01, 5.89556820e-02], [2.26021523e-01, 7.01343504e-02, 4.07186700e-02, ..., 3.29680917e-01, 2.40345734e-01, 1.94683189e-02], ..., [4.90731079e-01, 6.65312479e-01, 7.90649553e-01, ..., 8.49850498e-01, 8.47466322e-01, 3.01141839e-01], [1.32789591e-01, 3.01567641e-01, 9.82754771e-01, ..., 3.83435223e-02, 7.55896741e-01, 7.44475982e-01], [7.13453261e-01, 5.70347081e-02, 1.07477961e-01, ..., 4.77604550e-01, 8.05875931e-01, 6.39158677e-01]]])
- created_at :
- 2021-11-14T11:51:53.110470
- arviz_version :
- 0.11.4
<xarray.Dataset> Dimensions: (chain: 10, draw: 1000, dim_1: 10000) Coordinates: * chain (chain) int64 0 1 2 3 4 5 6 7 8 9 * draw (draw) int64 0 1 2 3 4 5 6 7 8 ... 992 993 994 995 996 997 998 999 * dim_1 (dim_1) int64 0 1 2 3 4 5 6 ... 9993 9994 9995 9996 9997 9998 9999 Data variables: x (chain, draw, dim_1) float64 0.3969 0.9378 0.349 ... 0.8059 0.6392 Attributes: created_at: 2021-11-14T11:51:53.110470 arviz_version: 0.11.4xarray.Dataset
arviz.ess#
%%time
%%memit
az.ess(idata_numpy)
peak memory: 1034.65 MiB, increment: 10.89 MiB
CPU times: user 21 s, sys: 192 ms, total: 21.2 s
Wall time: 21 s
Tip
Set the most common default dask_kwargs when enabling Dask in order to simplify
future function calls. If needed, those default kwargs can always be overrriden
with the function specific dask_kwargs argument.
Dask.enable_dask(dask_kwargs={"dask": "parallelized", "output_dtypes": [float]})
%%time
%%memit
ess = az.ess(idata_dask)
with ResourceProfiler(dt=0.25) as rprof:
ess.compute()
peak memory: 1035.45 MiB, increment: 0.79 MiB
CPU times: user 643 ms, sys: 104 ms, total: 747 ms
Wall time: 15.8 s
Each chunk also contains the evaluation expression which will be calculated in parallel and on-the-fly
ess.data_vars["x"].data.visualize()
rprof.visualize()
Dask.disable_dask()
Here, dask enabled method consumed around ~400MB memory which is around ~360MB lesser than the vanilla method (also considering the memory consumption of the Numpy Array ). Dask enabled method is also a bit faster.
arviz.rhat #
%%time
%%memit
az.rhat(idata_numpy)
peak memory: 1035.81 MiB, increment: 0.30 MiB
CPU times: user 32.7 s, sys: 167 ms, total: 32.9 s
Wall time: 32.5 s
Dask.enable_dask(dask_kwargs={"dask": "parallelized", "output_dtypes": [int]})
We have now enabled Dask with incorrect default kwargs, which we have to override in the function call:
%%time
%%memit
rhat = az.rhat(idata_dask, dask_kwargs={"output_dtypes": [float]})
with ResourceProfiler(dt=0.25) as rprof:
rhat.compute()
peak memory: 1036.70 MiB, increment: 0.88 MiB
CPU times: user 709 ms, sys: 156 ms, total: 865 ms
Wall time: 20.6 s
rprof.visualize()
Dask.disable_dask()
arviz.hdi#
%%time
%%memit
az.hdi(idata_numpy, hdi_prob=0.68)
peak memory: 1037.05 MiB, increment: 0.20 MiB
CPU times: user 5.9 s, sys: 63.8 ms, total: 5.96 s
Wall time: 5.95 s
Dask.enable_dask(dask_kwargs={"dask": "parallelized", "output_dtypes": [float]})
With arviz.hdi() we are introducing a new dimension to the output, the one containing the lower and higher HDI limits, so we need to use dask_gufunc_kwargs from xarray.apply_ufunc() which is passed as **kwargs first to arviz.wrap_xarray_ufunc(), then to xarray.apply_ufunc().
%%time
%%memit
hdi = az.hdi(idata_dask, hdi_prob=0.68, dask_gufunc_kwargs={"output_sizes": {"hdi": 2}})
with ResourceProfiler(dt=0.25) as rprof:
hdi.compute()
peak memory: 1037.55 MiB, increment: 0.50 MiB
CPU times: user 266 ms, sys: 78.1 ms, total: 344 ms
Wall time: 2.78 s
rprof.visualize()
Dask.disable_dask()
client.close()
In all the examples, it’s noticeable that:
Data structures provided by dask reduces the overall memory footprint, as it divides them into multiple chunks.
Due to the breakdown of complex computations into small tasks and parallelizing the executions, dask supported methods achieve significant performance gain.
